Streamlining Workflows
Streamlining Workflows
This framework allows you to design and implement complex pipelines, and deploy them on your institution’s computing cluster. This has been built keeping in mind the needs of bioinformatics workflows. However, it is easily extendable to any field where a series of steps (shell commands) are to be executed in a (work)flow to process big data.
Highlights
- No new syntax or language. Put all shell commands as a tsv file called flow mat.
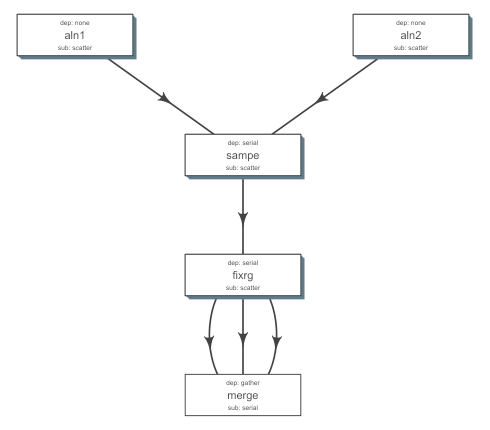
- Define the flow of steps using a simple tsv file (serial, scatter, gather, burst…) called flow def.
- Works on your laptop/server or cluster (/cloud).
- Supports multiple cluster computing platforms (torque, lsf, sge, slurm …), cloud (star cluster) OR a local machine.
- One line installation (
install.packages("flowr")) - Reproducible and transparent, with cleanly structured execution logs
- Track and re-run flows
- Lean and Portable, with easy installation
- Fine grain control over resources (CPU, memory, walltime of each step.
- Manage tools and default options using a companion params package.
- Access all R function from cmd line using funr.
Disclamer: Since we are using the same source for HTML and PDF, some plots/tables may not render perfectly in the PDF.
A few lines, to get started
## Latest stable release from CRAN (updated every other month)
## visit docs.flowr.space/install for more details
## for a latest official version (from CRAN)
Rscript -e 'install.packages("flowr", repos = c(CRAN="http://cran.rstudio.com"))'
## Latest stable release from DRAT (updated every other week); CRAN for dependencies
Rscript -e 'install.packages("flowr", repos = c(CRAN="http://cran.rstudio.com", DRAT="http://sahilseth.github.io/drat"))'
Rscript -e 'library(flowr);setup()'
# Run an example pipeline
# style 1: sleep_pipe() function creates system cmds
flowr run x=sleep_pipe platform=local execute=TRUE
# style 2: we start with a tsv of system cmds
# get example files
wget --no-check-certificate http://raw.githubusercontent.com/sahilseth/flowr/master/inst/pipelines/sleep_pipe.tsv
wget --no-check-certificate http://raw.githubusercontent.com/sahilseth/flowr/master/inst/pipelines/sleep_pipe.def
# submit to local machine
flowr to_flow x=sleep_pipe.tsv def=sleep_pipe.def platform=local execute=TRUE
# submit to local LSF cluster
flowr to_flow x=sleep_pipe.tsv def=sleep_pipe.def platform=lsf execute=TRUEResources
- For a quick overview, you may browse through, these introductory slides.
- The overview provides additional details regarding the ideas and concepts used in flowr
- We have a tutorial which can walk you through creating a simple pipeline.
- Or a tutorial explaining pipeline for fastq to bam.
- Additionally, a subset of important functions are described in the package reference page
- You may follow detailed instructions on installing and configuring
- You can use flow creator, a shiny app to aid in designing a shiny new flow. This provides a good example of the concepts
Troubleshooting
- Refer to the advanced configuration section or post issues on github’s issues page.
Talks/Slides
- Updated introduction (Sep, 2015)
- An earlier introduction at MD Anderson Comp. Biology meeting (July, 2015)
Acknowledgements
- Andy Futreal
- Jianhua Zhang
- Samir Amin
- Roger Moye
- Kadir Akdemir
- Ethan Mao
- Henry Song
- An excellent resource for writing your own R packages: r-pkgs.had.co.nz